
Probabilistic models for networks
You will find here all the material I use for my 3 hours class
on probabilistic models for network analysis.
Introduction
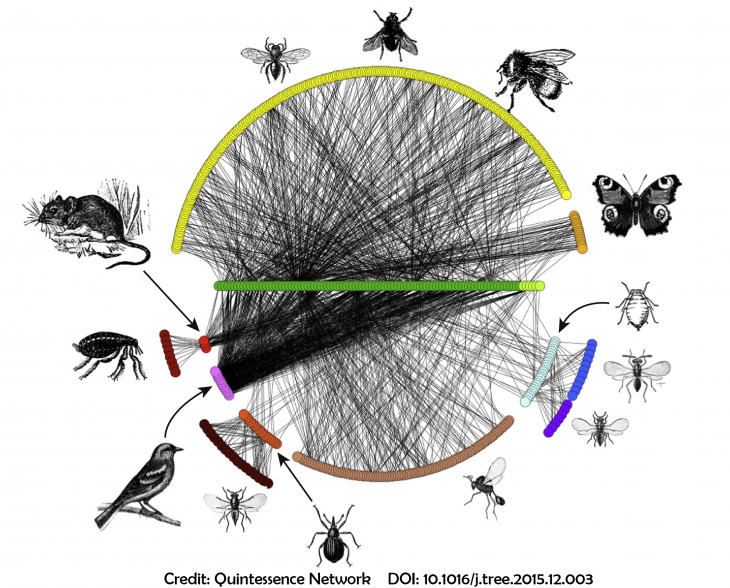
Ecological networks are made up of nodes, representing biological entities of interest and edges representing the interaction being studied. Stochastic block models (SBMs) and their extension to bipartite networks are convenient tools to modelize heterogeneity in (ecological) networks by introducing blocks of nodes sharing the same pattern of connection. This short-course presents SBMs for unipartite, bipartite or more complex networks and illustrates their flexibility.
Material for the theoretical part
Here are the slides of the course
- The first part is an Introduction to block models Part 1
- The second part will adress the Modelling of a collection of networks Part 2. This part is based on Chabert-Liddell, Barbillon, and Donnet (2022), available on arxiv. This second part will be dealt with only if we have time.
Additional material on Stochastic Block Models can be found the book Chapter 6 Using Latent Block Models to Detect Structure in Ecological Networks in Aubert et al. (2022).
R-tutorial
This class will include an R-tutorial session available here. You can download the
TutorialBM.qmd file here
It is mainly based on the R-package sbm.
Please find informations here.
Some additional R packages are needed.
install.packages("sbm")
install.packages("GGally") # To plot networks
install.packages('network')
install.packages('RColorBrewer') # to have nice colors
install.packages('knitr') # to plot nice tablesNote that a shiny application is also proposed. You should install the last version on your machine
It can also be used online here (not always the latest version).
In case we have anough time, the second part of the tutorial will
deal with collection of networks and we will use the R package
colSBM available on Github.
The tutorial on colSBM is available at this
link
Data sets
- The first part of the tutorial will rely on the fungus-tree
interaction network studied by Vacher, Piou, and
Desprez-Loustau (2008),
available with the package
sbm.
- We may also analyze the multipartite ecological network collected by
Dáttilo et al. (2016) (also available in
sbm) - Finally, we will maybe use the dataset of Thompson and Townsend (2003) provided in the
colSBMpackage
library(sbm)
data("fungusTreeNetwork")
data("multipartiteEcologicalNetwork")
library(colSBM)
data("foodwebs")